Results on 4 downstream tasks
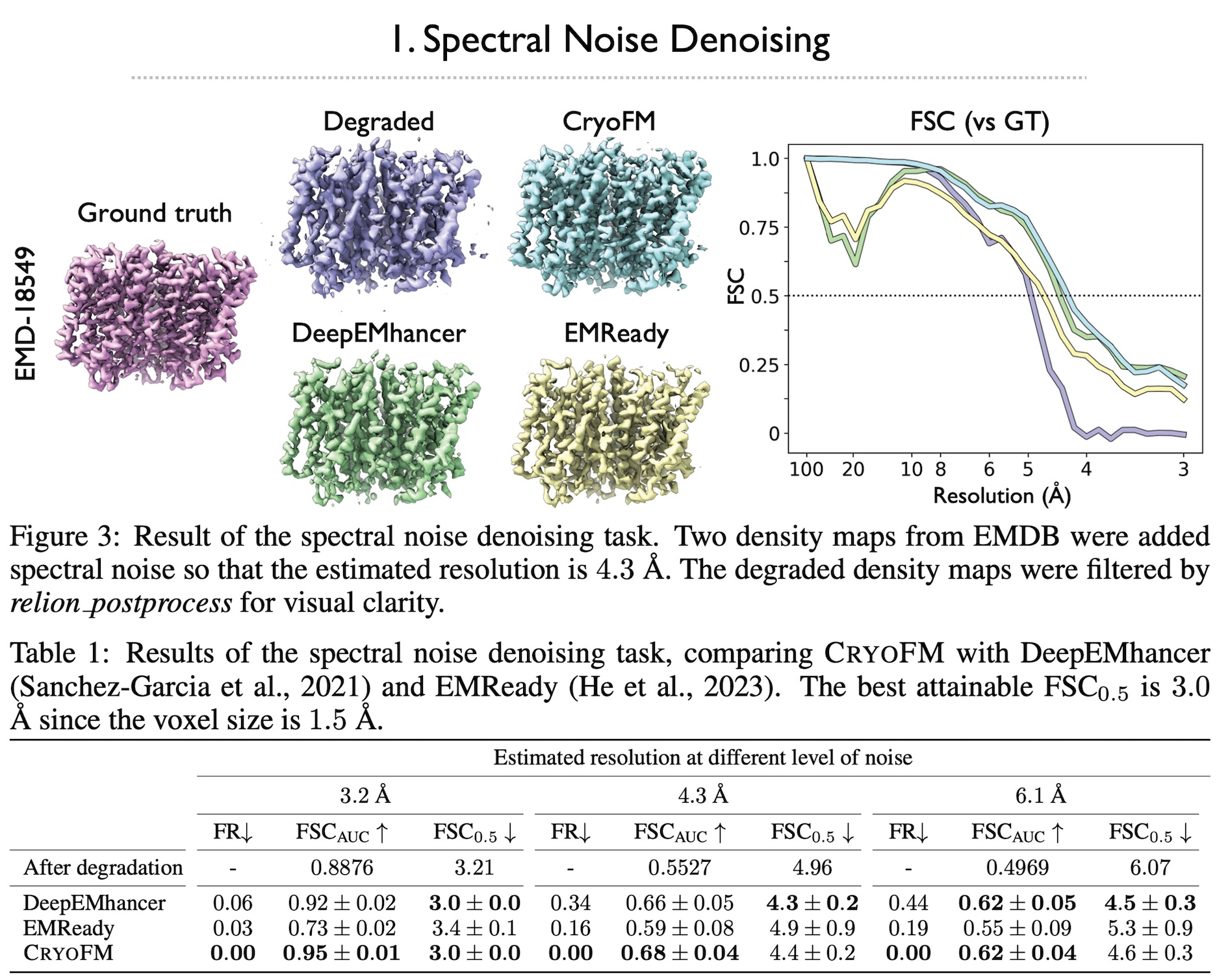
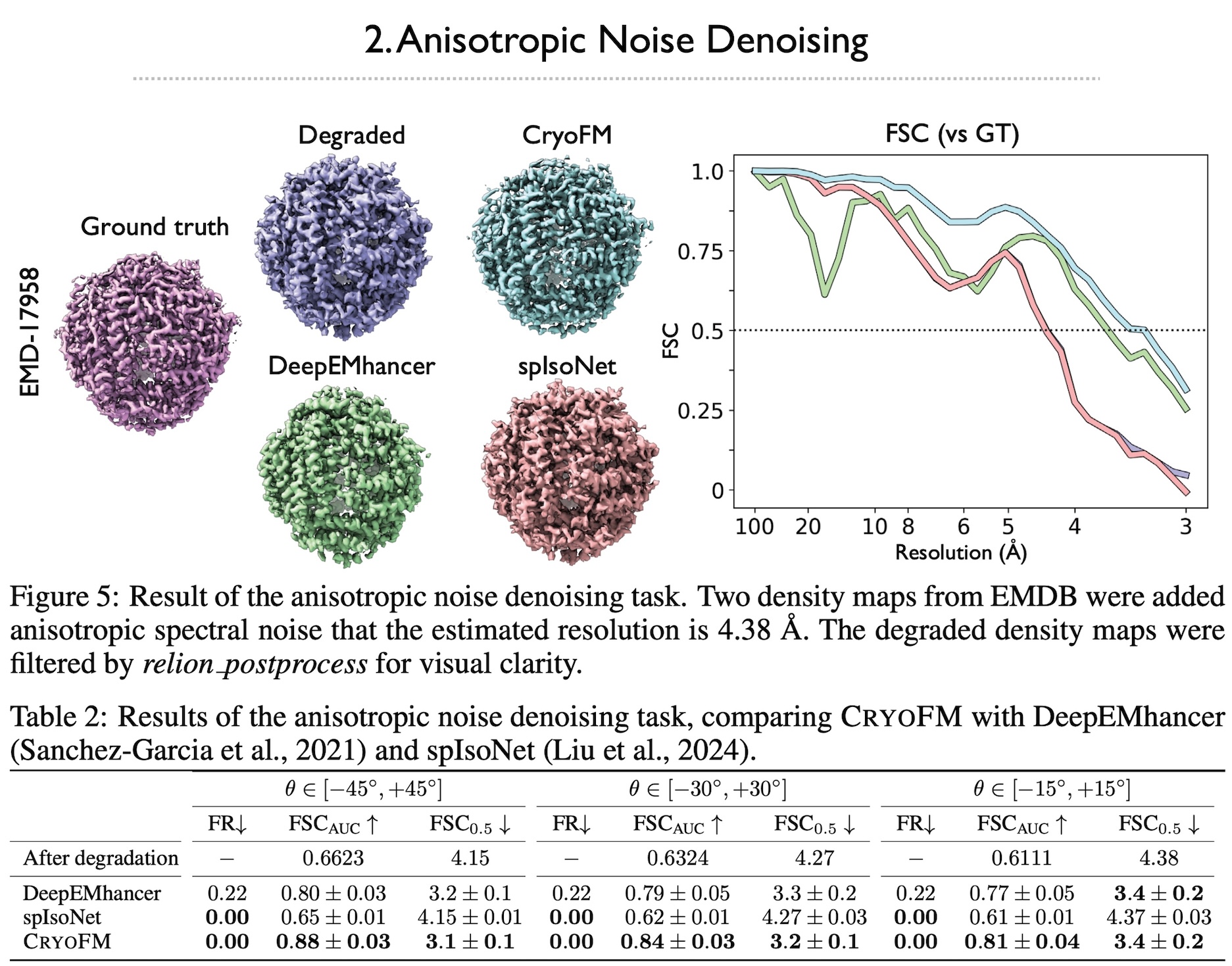
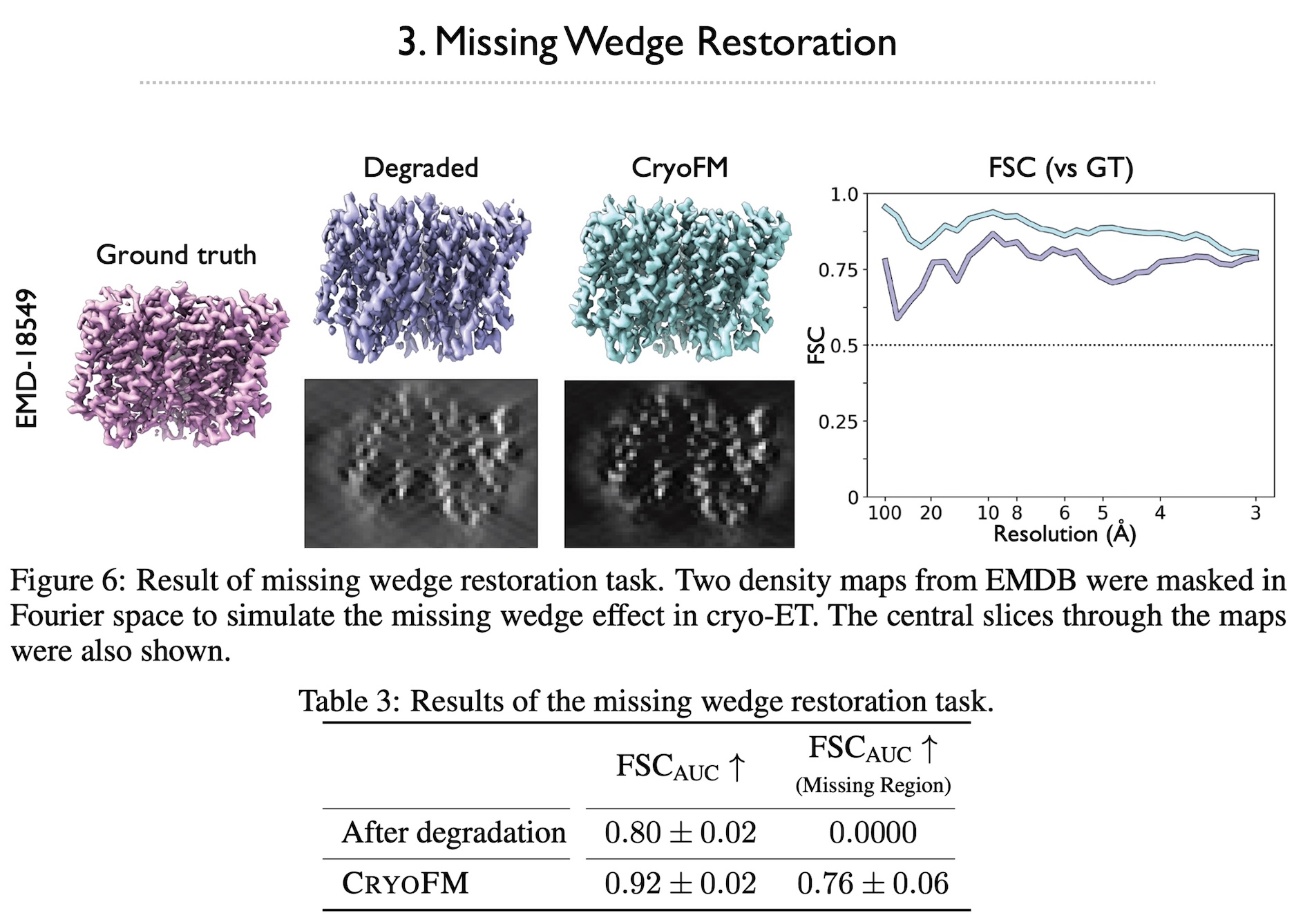
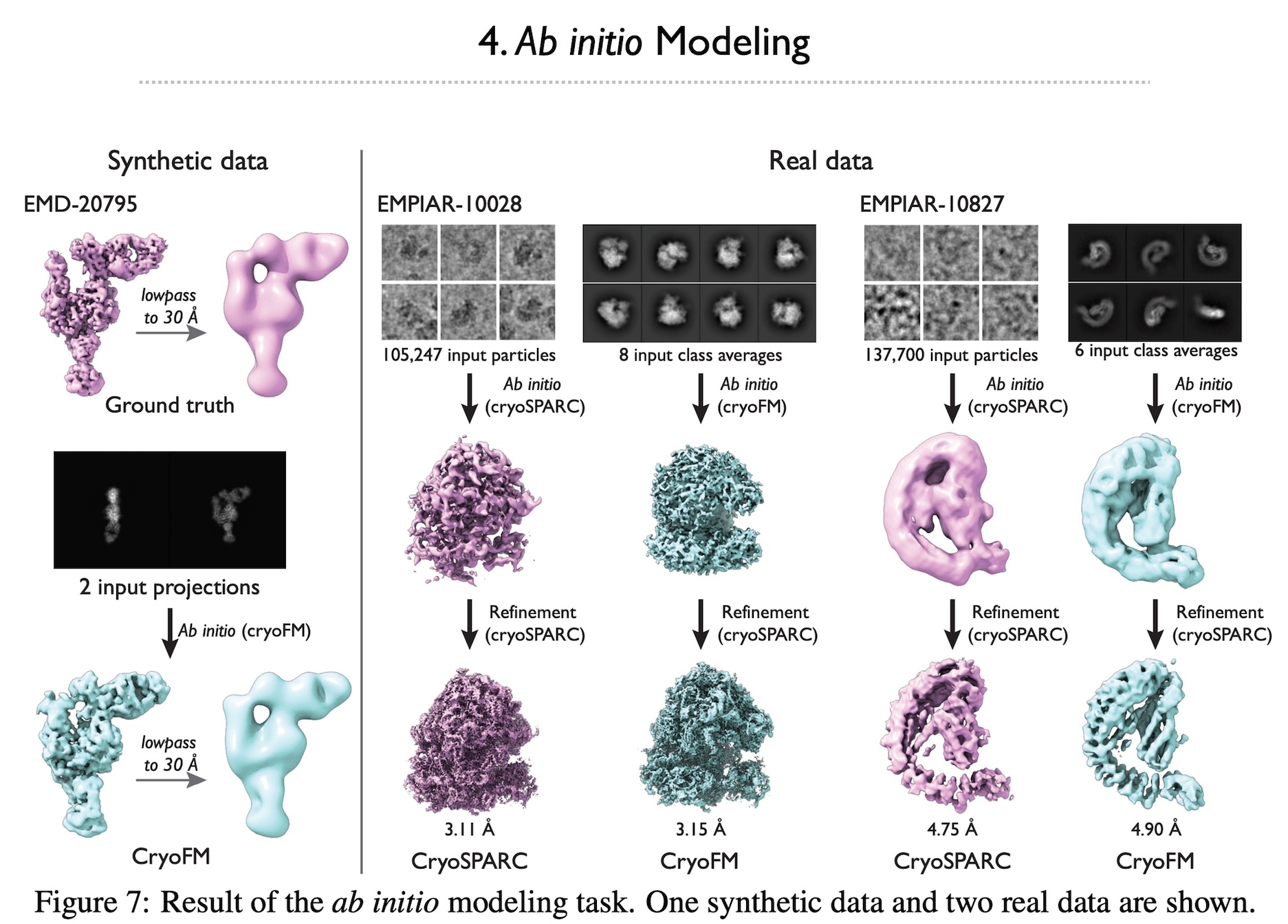
Cryo-electron microscopy (cryo-EM) is a powerful technique in structural biology and drug discovery, enabling the study of biomolecules at high resolution. Significant advancements by structural biologists using cryo-EM have led to the production of over 38,626 protein density maps at various resolutions. However, cryo-EM data processing algorithms have yet to fully benefit from our knowledge of biomolecular density maps, with only a few recent models being data-driven but limited to specific tasks. In this study, we present cryoFM, a foundation model designed as a generative model, learning the distribution of high-quality density maps and generalizing effectively to downstream tasks. Built on flow matching, cryoFM is trained to accurately capture the prior distribution of biomolecular density maps. Furthermore, we introduce a flow posterior sampling method that leverages cryoFM as a flexible prior for several downstream tasks in cryo-EM and cryo-electron tomography (cryo-ET) without the need for fine-tuning, achieving state-of-the-art performance on most tasks and demonstrating its potential as a foundational model for broader applications in these fields.




@article {zhou2024cryofm,
title={CryoFM: A Flow-based Foundation Model for Cryo-EM Densities},
author={Yi Zhou and Yilai Li and Jing Yuan and Quanquan Gu},
year={2024},
eprint={2410.08631},
archivePrefix={arXiv},
primaryClass={q-bio.BM},
url={https://arxiv.org/abs/2410.08631},
}